For translational oncology, patient-derived xenograft (PDX) models offer a way to test novel drugs tested in real time on both the immunodeficient person and their matching PDX cohort. Created by implanting freshly resected, unmanipulated tumour fragments from a patient into mouse hosts, PDXs faithfully conserve the tumour’s three-dimensional histopathology, clonal heterogeneity, and molecular signatures shaped by the patient’s prior treatments. PDX is an industry standard when it comes to helping researchers with:
A 2023 review in Signal Transduction and Targeted Therapy called PDXs “an ideal choice in cancer treatment studies,” noting their superiority in recapitulating spatial structure and genomic features across disease stages. It is, to use a turn of phrase, the good stuff.
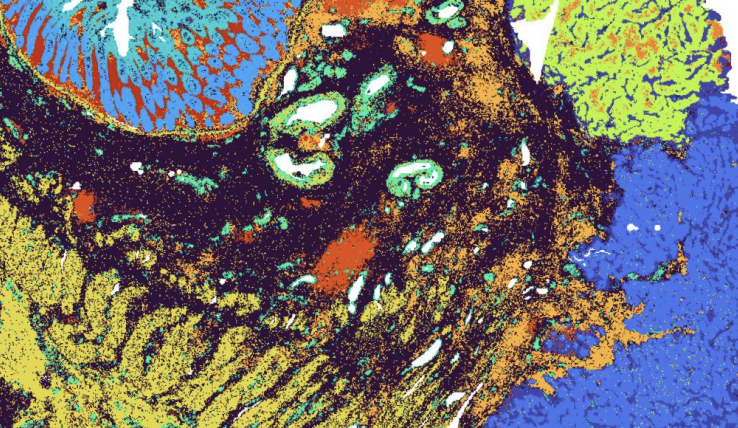
But PDX models are not without flaws. The same review — and other studies across the field — flag persistent hurdles when using PDX: loss of heterogeneity during passaging, selection bias, stromal replacement, and host-cell contamination that muddies genomic reads. Add the human-mouse noise headache and you quickly see why high-resolution spatial readouts like those achieved by 10x Genomics’ all-new Visium HD 3’ spatial assay are both essential and highly specialized.
Having an experienced service provider like BioChain support your spatial assay is one of the best ways to resolve your PDX problems and make running your study that much easier. Let’s take a closer look.
Bulk RNA-sequencing collapses thousands of cells into one average signal, masking border-zone biology where resistance often incubates. Even standard spatial assays (55 µm spots) merge five to ten cells per barcode — better, but still blurry. Here’s a breakdown of the problem:
The very artefacts and evolutionary dynamics that critics flag as limitations of PDX models (clonal drift, stromal replacement, species contamination) are precisely the features bulk RNA-seq is least equipped to resolve. What researchers using PDX really need is a way to:
Traditional work-arounds — like laser-capture microdissection or serial single-cell dissociations — either sacrifice spatial context or introduce new batch effects. Moving to high-resolution, species-aware spatial assays lets you see those pitfalls, quantify them, and, when necessary, correct for them instead of unknowingly averaging them away.
High-definition slides are only as good as the sample journey that precedes them. That’s where BioChain’s spatial biology services close the loop:
Bottom line: you supply the xenograft slides, we return a cell-level map of tumour–stroma interactions.PDX models remain oncology’s translational darling, but the field has outgrown mosaic-level views. Visium HD 3′ finally lets researchers watch human tumour evolution within a living mouse micro-environment, one cell at a time. Couple that capability with BioChain’s decades-deep expertise, ISO-certified rigour, and full-stack spatial platform menu, and you have a turnkey path from scarce xenograft tissue to publishable, decision-ready insights.
___
Ready to sharpen your PDX vision?
Chat with a BioChain spatial scientist today to learn how Visium HD 3′ plus BioChain can accelerate your next oncology breakthrough.